Research
We are aiming to optimize and apply CRISPR-mediated genome engineering technology to develop efficient gene therapy for muscular disorders in human and improve meat quality in livestock.
To achieve these goals, we are currently focusing on the following projects:
Develop in vivo delivery shuttles for genome editing tools
The evolving of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas (CRISPR-associated protein) system holds great promise of achieving precise editing in the genome with simplicity and versatility, making gene therapy more feasible for human diseases. How to deliver CRISPR-Cas system into the correct tissues and cell types in a safe and efficient way is perhaps the biggest barrier to fulfill its potential in biomedical and agriculture fields. We are proposing to genetically engineer exosomes, nanosized natural intercellular delivery vehicles, as shuttle for CRISPR-Cas system to achieve cell tropism-dependent targeted genome editing (Fig. 1). In addition, taking advantage of the ability of exosomes to penetrate the blood brain barrier (BBB), it is promising to send CRISPR mediated gene therapy into center nerve system (CNS) for neurological disorders.
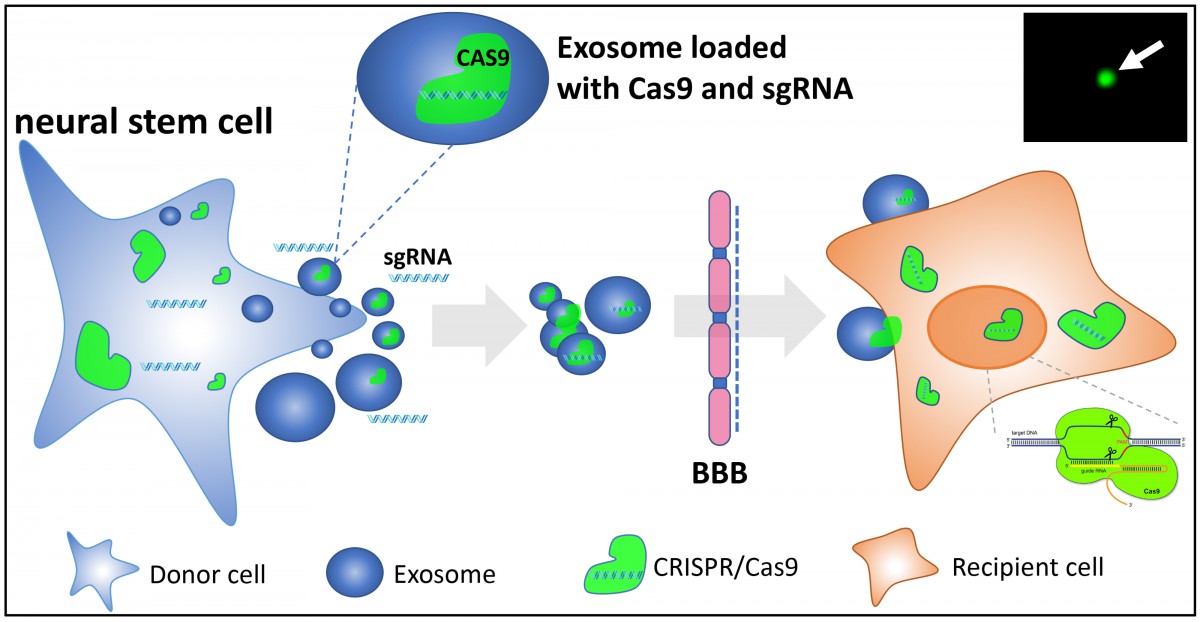
Figure 1: A schematic view of engineering NSC-derived exosomes loaded with CRISPR-Cas9 machinery as a gene therapy tool. The arrow in the inset points to a fluorescent exosome, indicating successful loading of Cas9-GFP into exosome. (Yao et al., unpublished)
Develop gene therapies for neuromuscular dystrophies (Amyotrophic lateral sclerosis (ALS))
ALS is the most frequently occurring, fatal neuromuscular degenerative disease. This devastating disease affects upper and lower motor neurons from the brain and spinal, causing skeleton muscle weakness and atrophy, eventually leads to paralysis and death (average survival time is 3-5 years). However, no effective treatment so far. About 90% of ALS cases are sporadic, while 10% are familial due to genetic mutation, and the most common associated gene is C9ORF72. We are developing CRISPR mediated gene therapy to treat the genetic ALS caused by hexanucleotide repeats in the first intron of C9ORF72 gene. These expanded DNA repeats cause loss of function of C9ORF72 by partially transcription inhibition. More importantly, gain-of-toxicity from the aberrant RNAs derived from the DNA repeats forming these intranuclear foci and dipeptides proteins translated from these repeats containing RNAs account for the majority of the pathogenic phenotypes (Fig. 2). We are establishing induced pluripotent stem cells (iPSCs) and animal disease models of C9ALS to evaluate potential therapy and screen for novel drug targets.
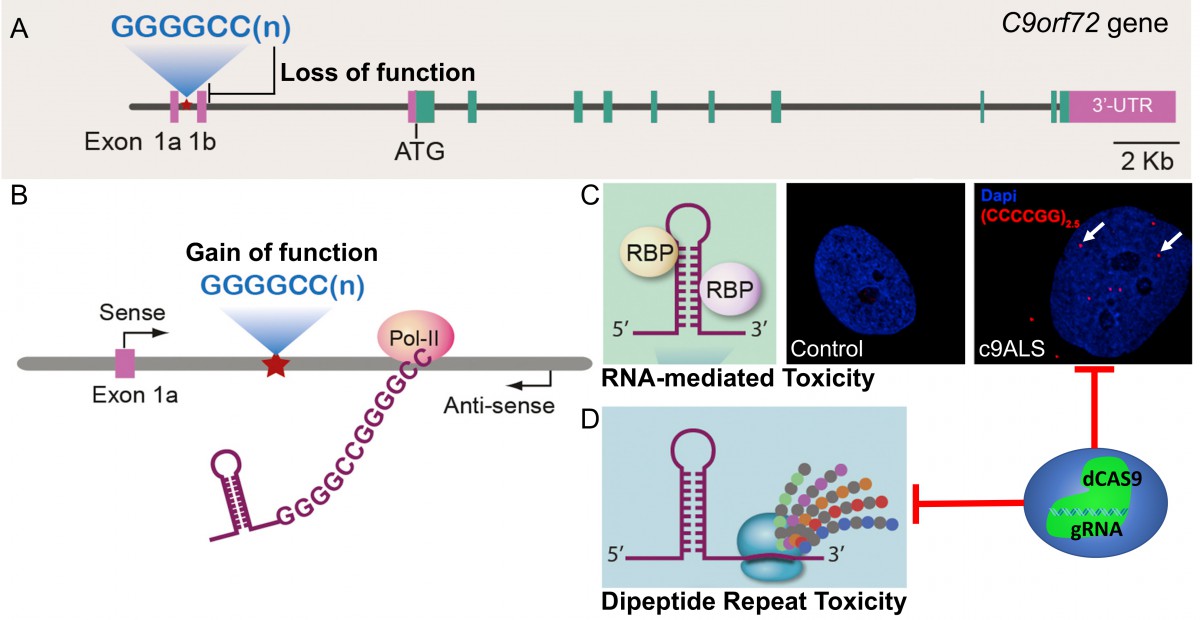
Figure 2: Proposed disease mechanisms and pathological phenotypes of c9ALS (A) haploinsufficiency; (B-D): Gain of toxicities by RNA molecules (C) and dipeptides repeats protein (D) from the hexanucleotide expansions. The arrows in (C) point to the pathologic RNA foci in c9ALS cells. RBP: RNA binding protein. (Adapted from Gitler et al., 2016; Yao et al., unpublished)
Understand the role of Fibro/adipogenic progenitors (FAPs) at molecular level in response to different muscle damages
Skeletal muscle is capable of regeneration in response to different damages. During this process, FAPs function as a double-edged sword. FAPs can either exert a positive influence on regeneration by providing tropic factors to support satellite cell differentiation, or trigger fibrosis and fatty degeneration after differentiating into fibroblast and adipocytes. Interestingly, FAPs activate pathogenic STAT3-IL-6 signaling in skeletal muscle denervation, which promotes muscle atrophy. The functional versatility of FAPs in response to different muscle injuries makes it a key effector and therapeutic target for muscular dystrophies. However, the signals regulating their growth, survival and differentiation remain largely unknown. We will apply high throughput methods (RNA-seq, ChIP-seq, ATAC-seq) to analyze the transcriptome, enhancer activity in FAPs from different muscular disease models (ALS, Duchenne Muscular Dystrophy and cellular models induced by chemicals) to uncover key factors/pathways as novel targets of gene therapies for muscular dystrophies.
Investigate gene regulatory network in FAPs involved intramuscular adipocyte development
FAPs originally differentiated from mesenchymal stem cells, are the major source of intramuscular adipocytes for marbling fat accumulation in livestock. We are aiming to improve meat quality through enhancing intramuscular adipogenesis and reducing the competitive fibrogenesis by manipulating FAP differentiation (Fig. 3). In this way, both the marbling and tenderness of meat will be increased since less fibrogenic differentiation reduces intramuscular connective tissues, thereby decreasing toughness of the meat. Specifically, we will 1) Identify enhancers of preadipogenic determinator Zfp423 in FAPs; 2) Uncover other key factors responsible for adipogenic commitment of FAPs; 3) Engineer FAPs through CRISPR-mediated genome editing to facilitate intramuscular adipogenesis in vitro and in vivo.
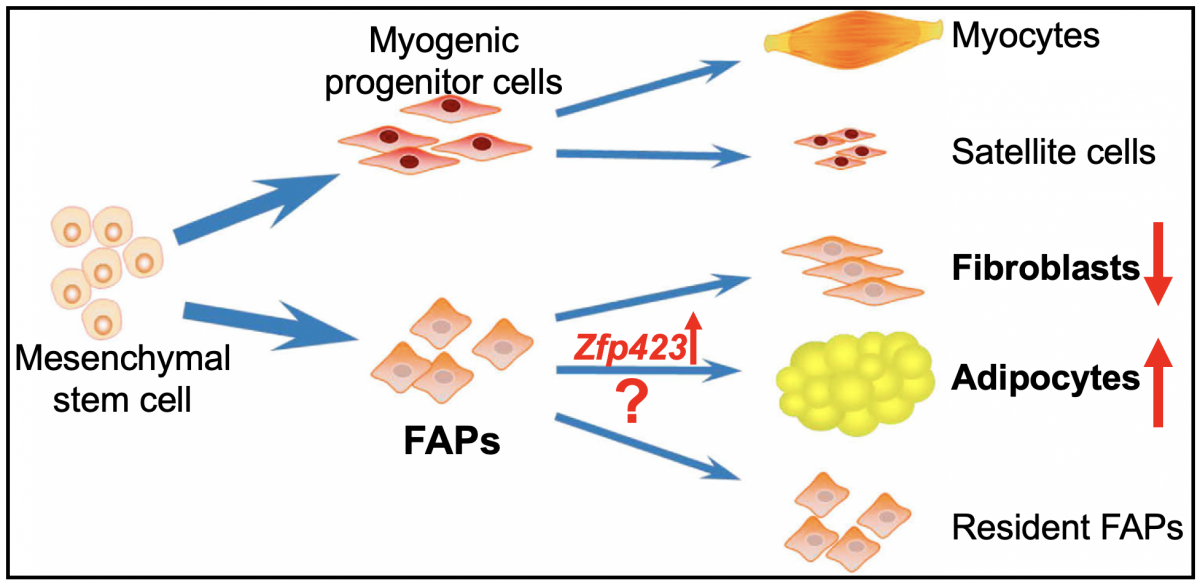
Figure 3: A schematic view of strategies to improve intramuscular adipocytes through increasing the expression of Zfp423 and other factors responsible for adipogenic commitment of FAPs. (Modified from Du et al., 2012)