Current Publications
Research Focus Areas
Mechanisms of plant immune action on bacteria.
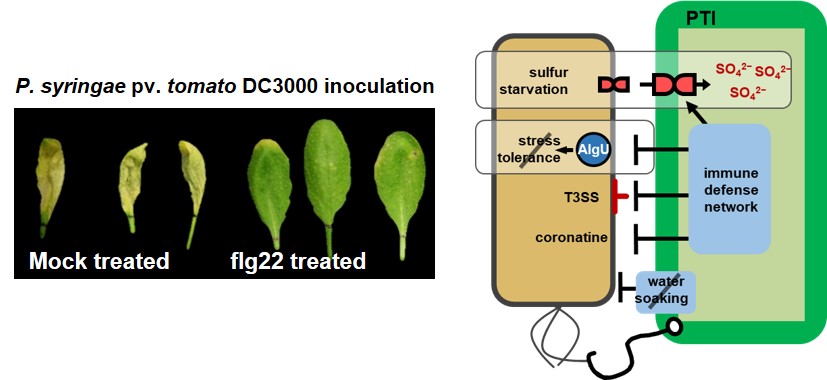
Like animal cells, plant cells deploy specialized transmembrane receptors (Pattern Recognition Receptors) with the capacity to recognize specific conserved molecular patterns of invading microbes (e.g. the flg22 epitope of bacterial flagellin). Recognition of microbial invaders can trigger a defensive immune response (Pattern Triggered Immunity, PTI) by plant cells that is able to inhibit disease caused otherwise competent pathogens. We are particularly focused on characterizing the mechanisms by which PTI suppresses bacterial proliferation and virulence. Using changes in patterns of bacterial gene expression during PTI as a guide, we are exploring the roles of sulfate sequestration and dysregulation of a conserved stress response pathway in plant immune action on bacteria.
The genetic and molecular basis of virulence for bacterial pathogens of important Georgia crops.
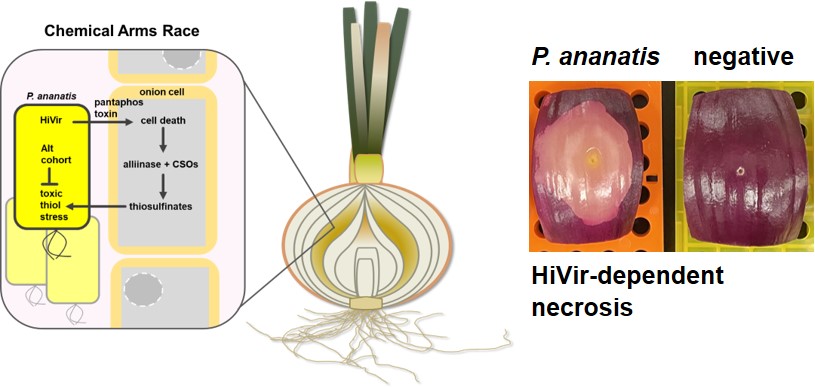
Onion: Onions are impacted by a wide diversity of plant pathogenic bacteria that routinely cause significant economic losses for Georgia Vidalia onion producers. We are characterizing the diverse mechanisms by which these bacterial pathogens exploit this major vegetable crop as a host. We have been focused on Pantoea onion pathogens, which couple production of phytotoxins with phytoanticipin tolerance to create an atypical framework for bacterial necrotrophy. In addition, we have been characterizing the genetic basis of Burkholderia gladioli onion virulence.
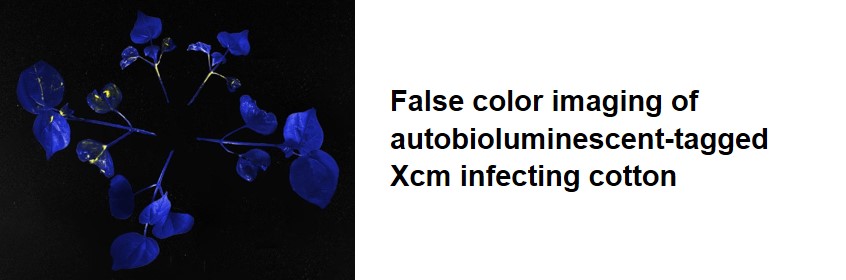
Cotton: We are characterizing the roles of seed-borne transmission and systemic movement of Xanthomonas citri pv. malvacearum in Cotton Bacterial Blight (CBB) and are working towards introducing new forms of CBB resistance into cotton using transgenic and genome editing approaches.